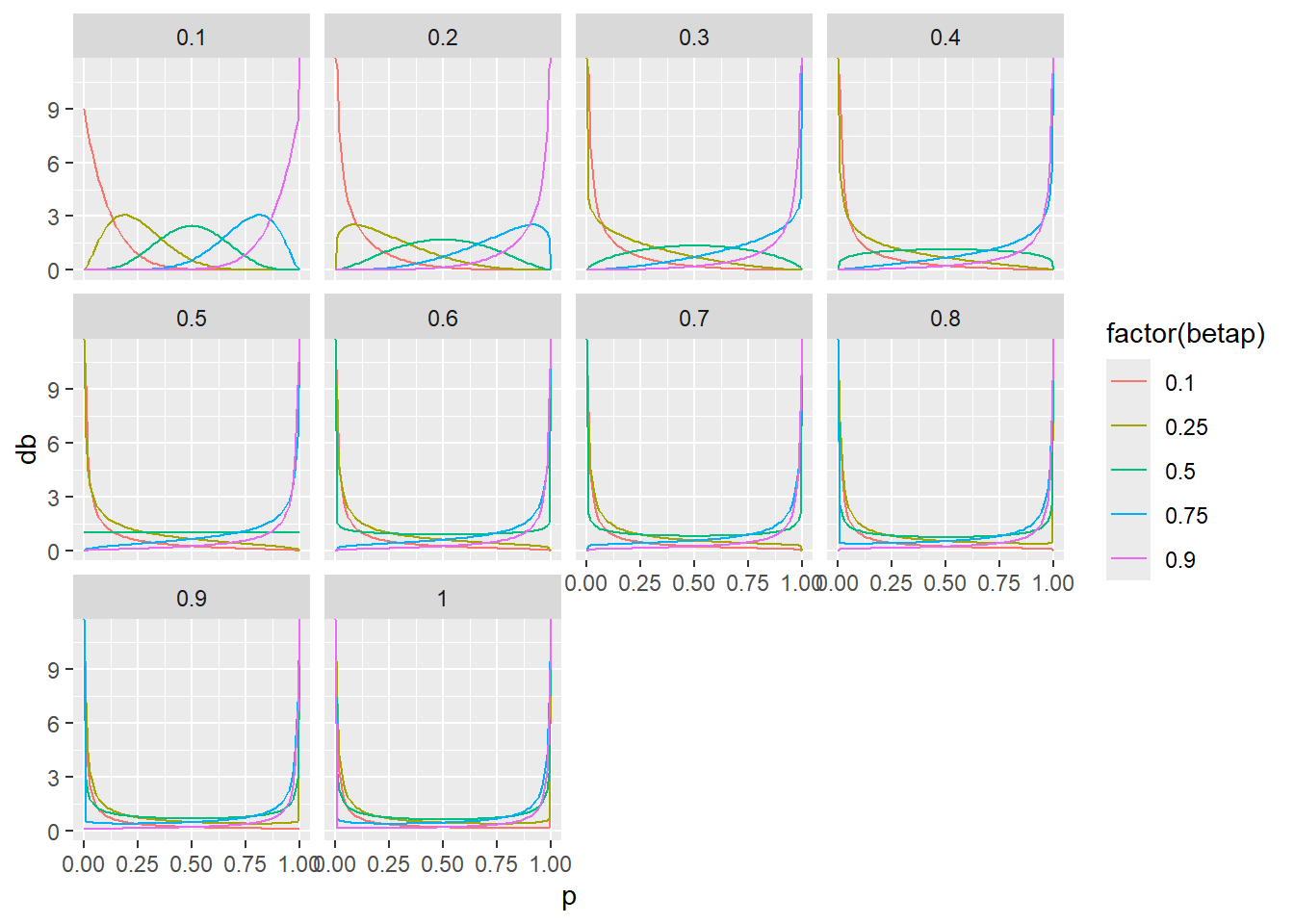
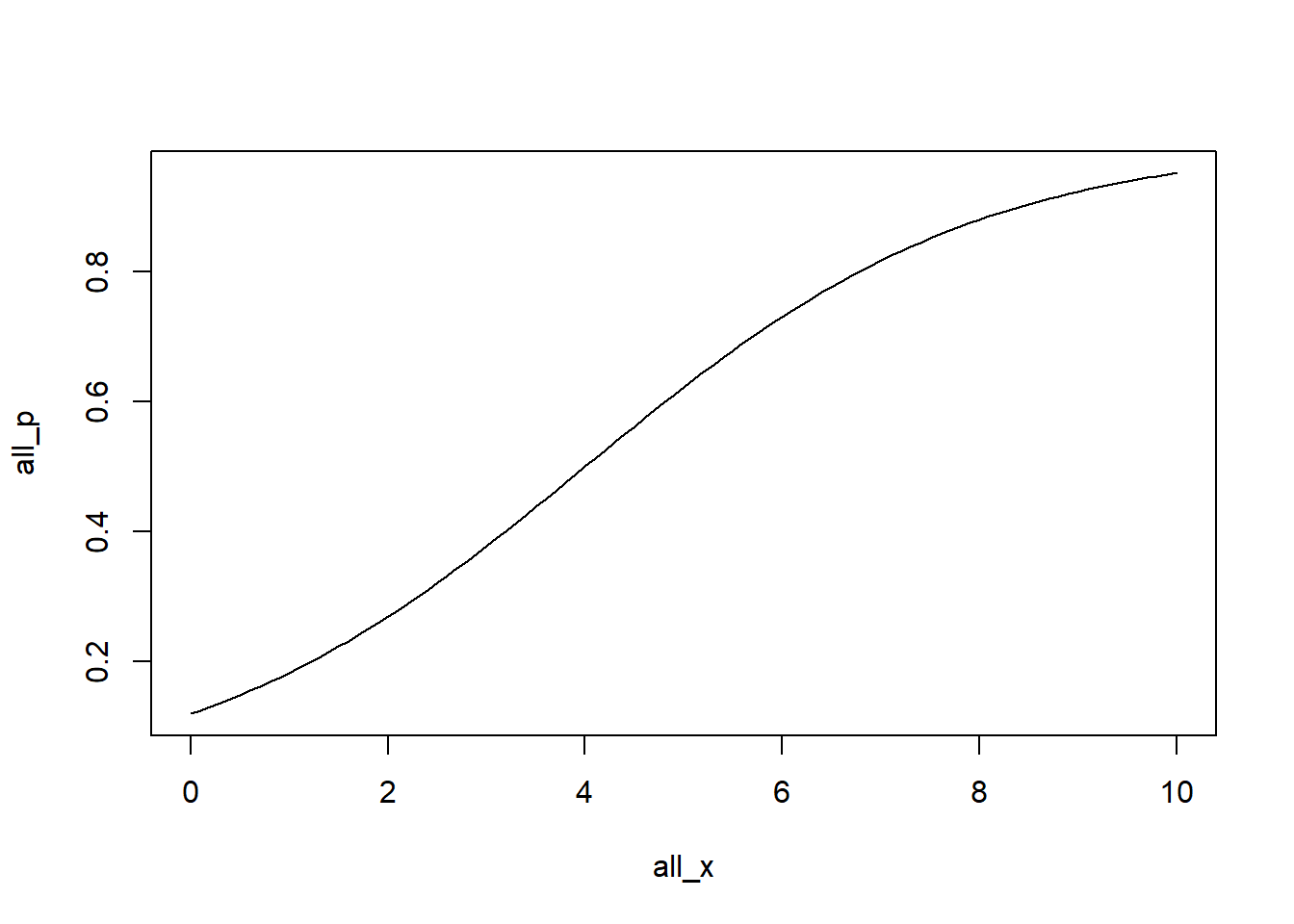This is fairly sketchy, just trying to see how the \(\phi\) from Harrison (2015 ) affect beta probability distributions.
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
<- seq (from = 0 , to = 1 , by = 0.01 )<- seq (from = 0.1 , to = 1 , by = 0.1 )
# ai <- p[1]/phi[1] # bi <- (1-p[1])/phi[1] # # db <- dbeta(p, ai, bi) <- tibble (p = NA , db = NA , betap = NA , phi = NA , .rows = 0 )for (j in 1 : length (phi)) {for (i in 1 : length (p)) {<- p[i]/ phi[j]<- (1 - p[i])/ phi[j]<- dbeta (p, ai, bi)<- tibble (p, db, betap = p[i], phi = phi[j])<- rbind (probdists, dbt)
ggplot (probdists |> filter (betap %in% c (0.1 , 0.25 , 0.5 , 0.75 , 0.9 )), aes (x = p, y = db, color = factor (betap))) + geom_line () + facet_wrap ("phi" )
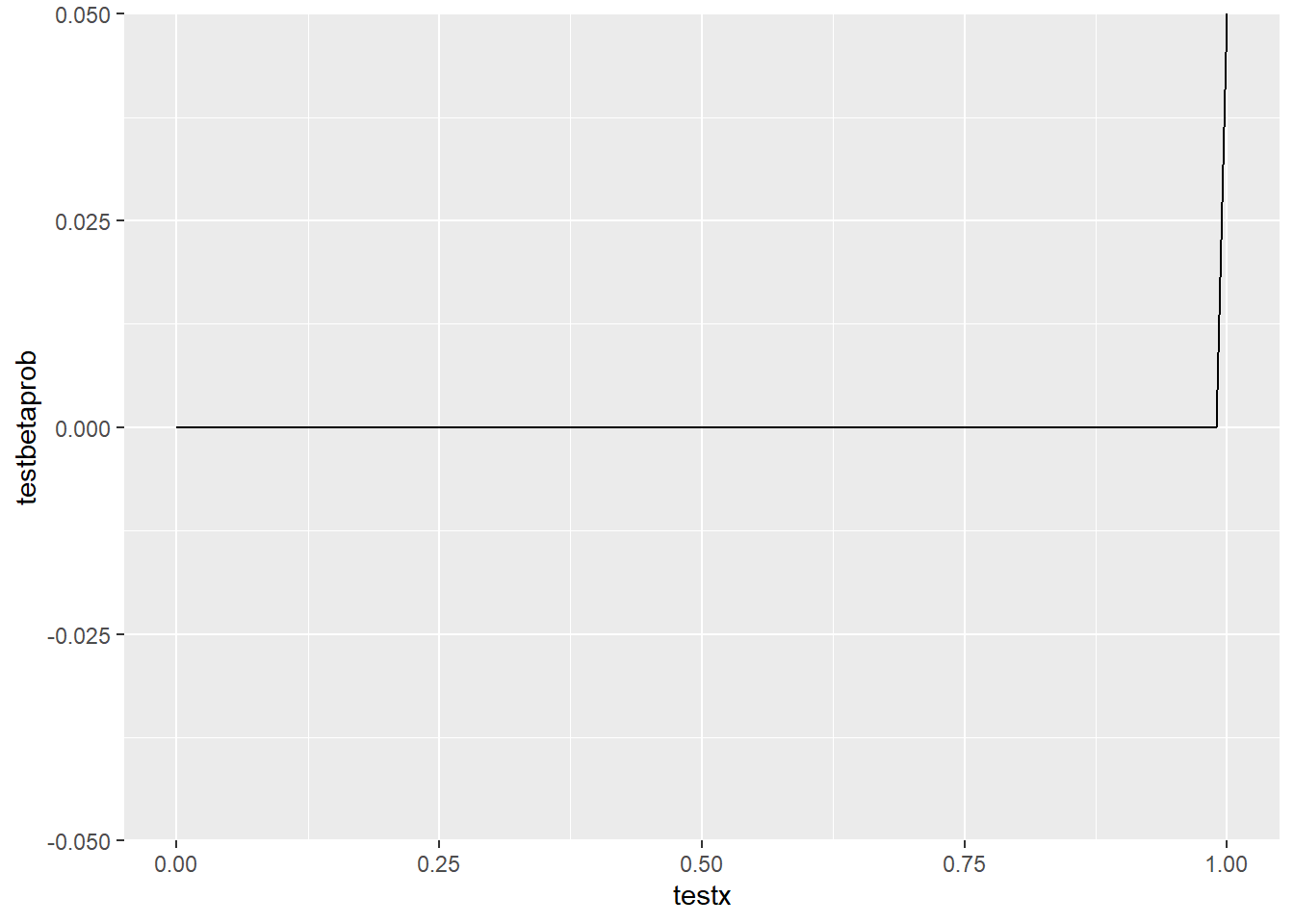
<- seq (from = 0 , to = 1 , by = 0.01 )<- dbeta (testx, ai, bi)<- tibble (testx, testbetaprob)ggplot (probdist, aes (x = testx, y = testbetaprob)) + geom_line ()
Random draws
<- - 2 <- 0.5 <- seq (0 , 10 , 0.1 )<- intercept + beta* all_x<- 1 / (1 + exp (- all_logit_p))
plot (all_x, all_p, type = 'l' )
References
Harrison, Xavier A. 2015.
“A Comparison of Observation-Level Random Effect and Beta-Binomial Models for Modelling Overdispersion in Binomial Data in Ecology & Evolution.” PeerJ 3 (July): e1114.
https://doi.org/10.7717/peerj.1114 .