knitr::opts_knit$set(root.dir = rprojroot::find_rstudio_root_file())Bom reference stations
library(ggplot2)
library(sf)Trying to find BOM gauge locations. Found reference stations. It’s a simple link, but have to use httr2 to download because there’s an error with the user_agent if we try to just download.file.
Mostly including this here as an example of changing user_agent.
http://www.bom.gov.au/waterdata/ has a clickable link to what I want, but the data is buried in a frame so can’t scrape.
The below is because I found a link to reference stations and wanted to see what they were.
Is there a url for BOM?
It’s just a csv, but have to faff about with httr2 and deparsing back to csv because need to pass a user agent or get a 403 error.
bom2 <- httr2::request("http://www.bom.gov.au/water/hrs/content/hrs_station_details.csv") |>
httr2::req_user_agent("md-werp") |>
httr2::req_perform() |>
httr2::resp_body_string() |>
readr::read_csv(skip = 11) |>
dplyr::select(site = `Station Name`,
gauge = `AWRC Station Number`,
owner = `Data Owner Name`,
Latitude, Longitude)Rows: 467 Columns: 8
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (5): AWRC Station Number, Station Name, Jurisdiction, Data Owner Name, D...
dbl (3): Latitude, Longitude, Catchment Area (km2)
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.basin <- read_sf(file.path('data','mdb_boundary', 'mdb_boundary.shp'))bom2 <- bom2 |>
# lat an long come in as chr because there is a line for 'undefined'
dplyr::filter(site != 'undefined') |>
st_as_sf(coords = c('Longitude', 'Latitude'), crs = 4326) |>
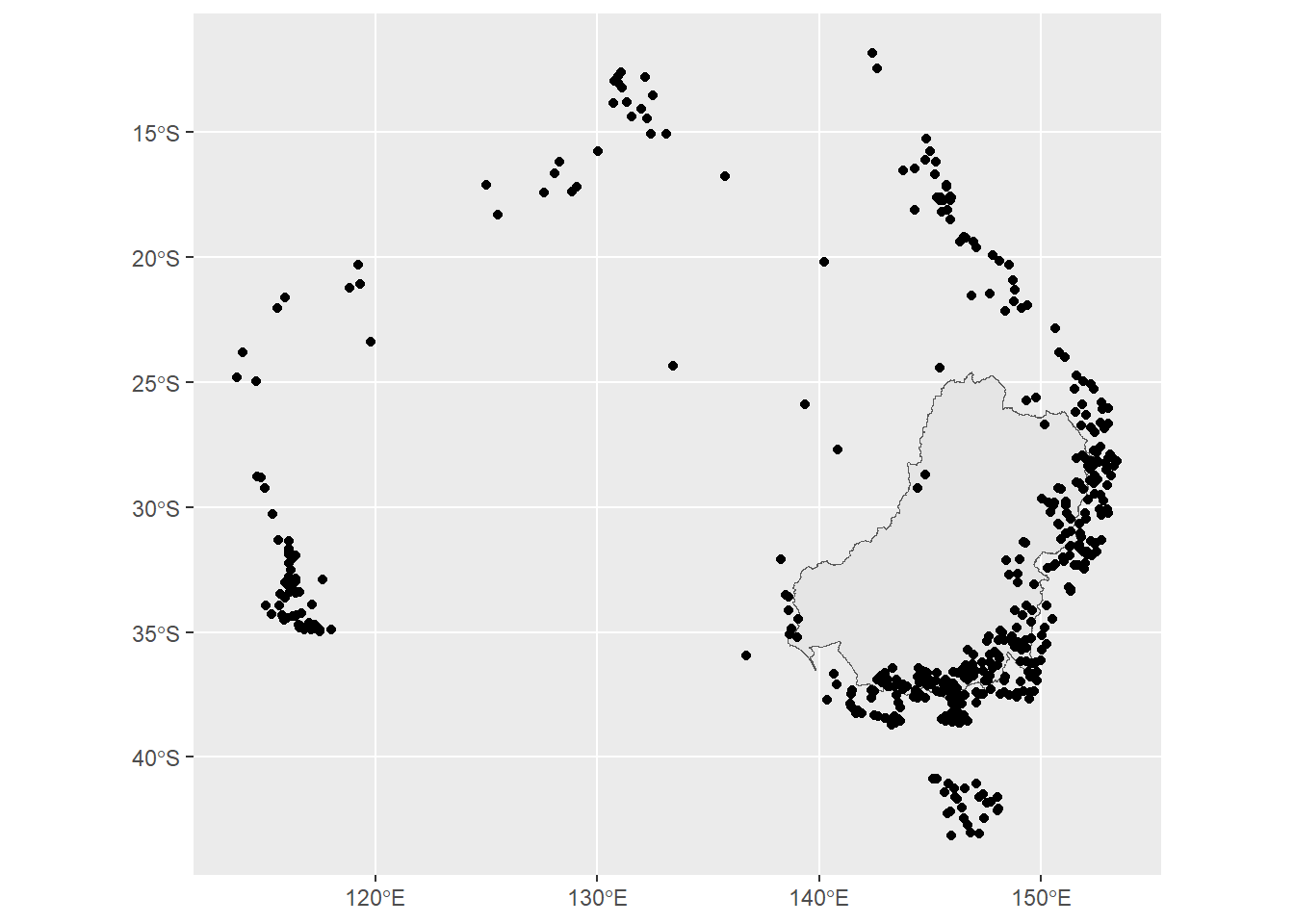
st_transform(crs = st_crs(basin))They’re not the gauges I’m looking for. Only 457, instead of 6500, and around the edges of the basin.
ggplot() +
geom_sf(data = basin) +
geom_sf(data = bom2)