library(ggforce)
library(tidyverse)
devtools::load_all()ℹ Loading galenRI need to create something like a Venn diagram, but scaled. None of the venn packages seem to do it. Can I get it to work with geom_circle and some math?
First, let’s say we have two sets of different size, with some overlap
If we want to just represent each set as a circle with area proportional to the number of items and not worrying about overlap, we can
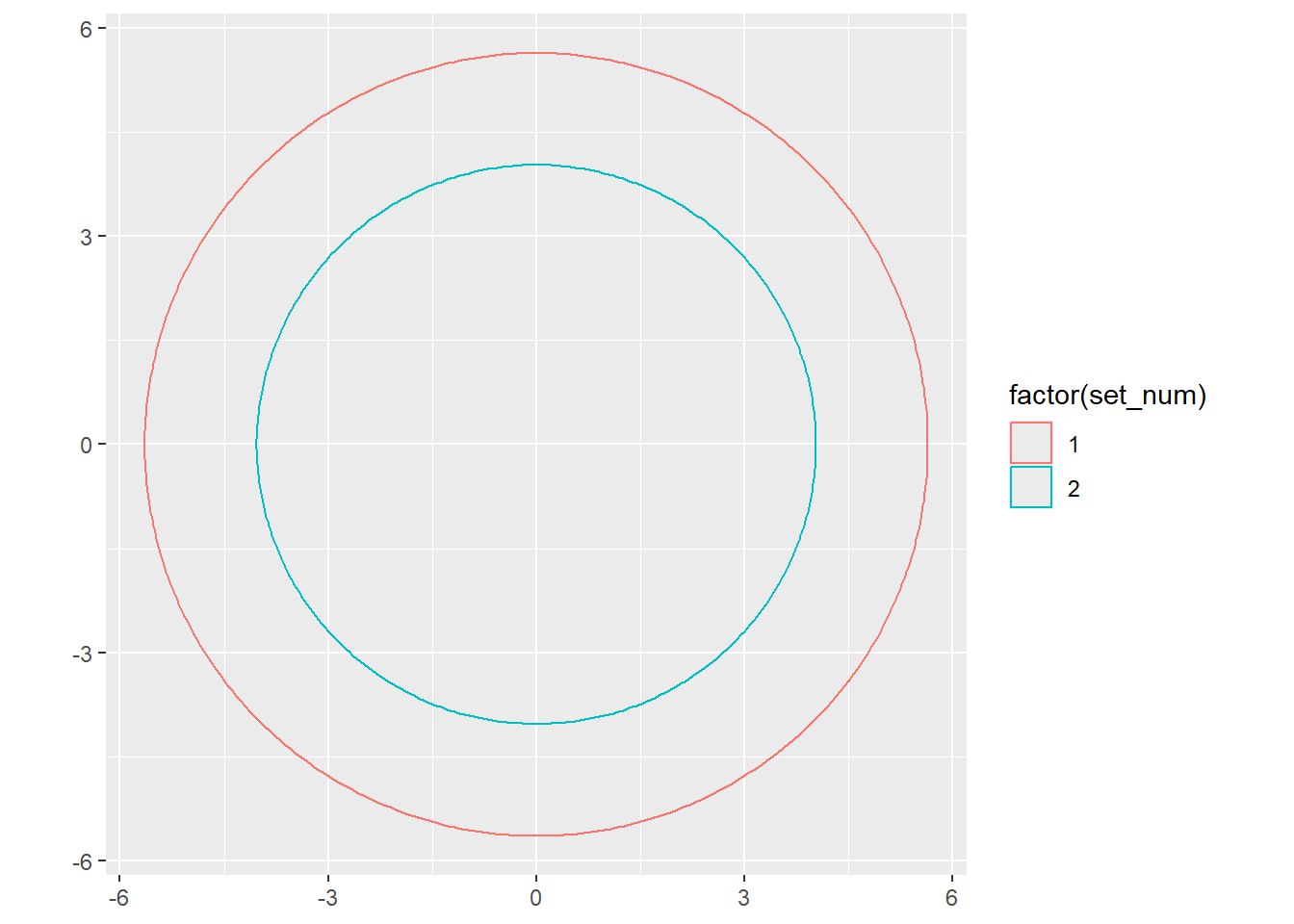
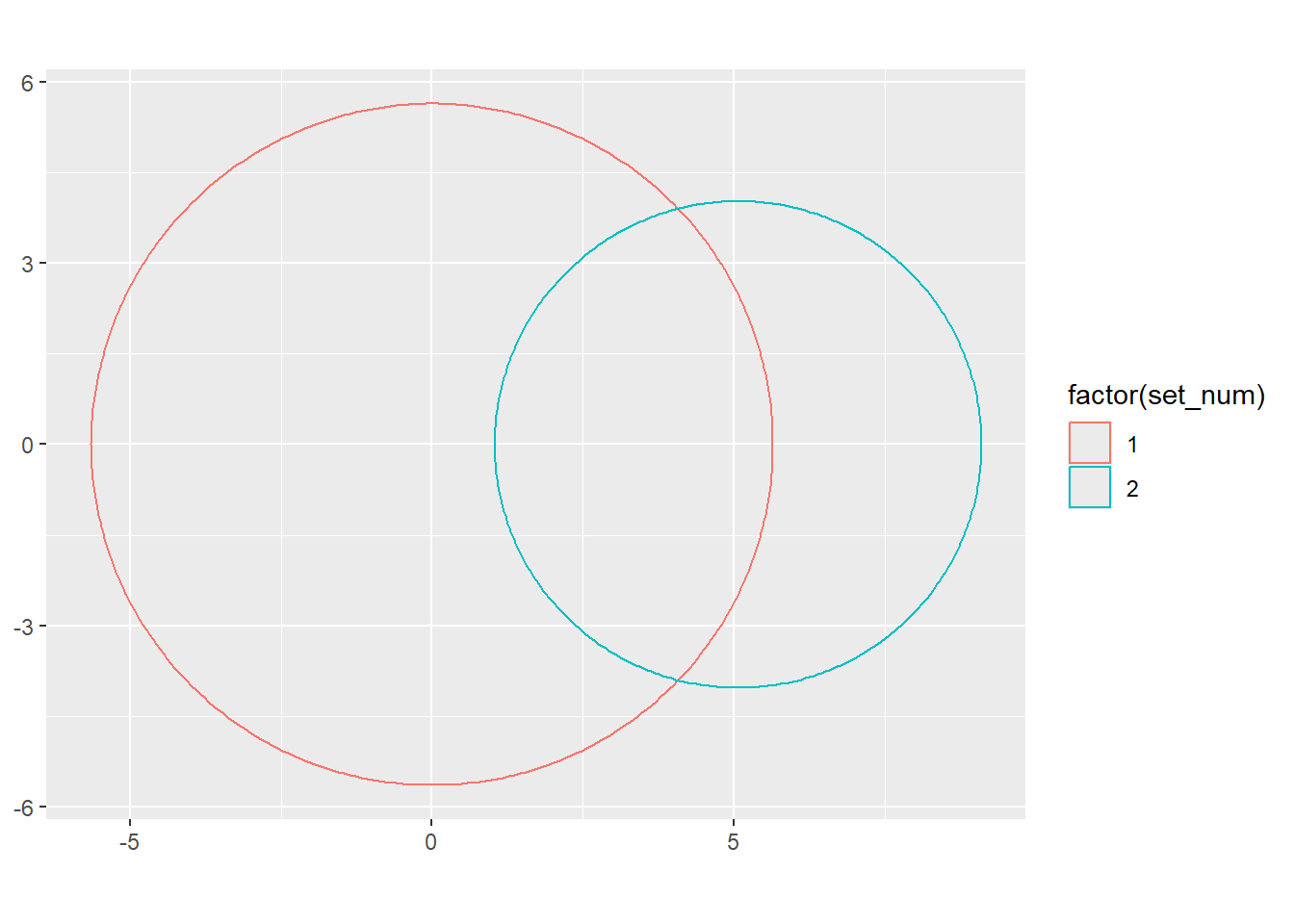
Now, though, I want an area of overlap of 25. So really, I need to find an offset for (let’s say) x0 for set2 that shifts that circle half in and half out.
Wolfram has a complicated formula (14) to get A (the area of the ‘lens’- the overlap) given the distance d between centers and the radii. Here, we have the area and radii, and want to solve for d. So can we do that?
A = r^2*acos((d^2 + r^2 - R^2) / (2*d*r)) +
R^2*acos((d^2 - r^2 + R^2) / (2*d*r)) -
0.5*sqrt((d+r-R) * (d-r+R) * (-d+r+r) * (d+r+R))According to Sage
pi - acos(-1/2*(R^2 - d^2 - r^2)/(d*r)) == 1/2*(2*(pi - acos(-1/2*(R^2 + d^2 - r^2)/(d*r)))*R^2 + 2*pi*r^2 - 2*A - sqrt(R^3*d + R^2*d^2 - R*d^3 - d^4 + (2*R - d)*r^3 - 2*r^4 + (2*R^2 + 3*R*d + 3*d^2)*r^2 - (2*R^3 + 3*R^2*d - d^3)*r))/r^2That’s not particularly helpful; d is not isolated.
I should be able to just do this numerically with optim or optimize, I think.
# calc_a <- function(d, radius1, radius2) {
# A <- radius1^2*acos((d^2 + radius1^2 - radius2^2) / (2*d*radius1)) +
# radius2^2*acos((d^2 - radius1^2 + radius2^2) / (2*d*radius1)) -
# 0.5*sqrt((d+radius1-radius2) * (d-radius1+radius2) * (-d+radius1+radius1) * (d+radius1+radius2))
#
# return(A)
# }
#
# opt_d <- function(d, area, radius1, radius2) {
#
# dcheck <- calc_a(d, radius1, radius2)
#
# return(dcheck - area)
# }# get the optimal shift
#opt_d and calc_a are defined in venn_distances.R
# find_d <- function(area, radii, radius1, radius2) {
# if (!missing(radii) & (!missing(radius1) | !missing(radius2))) {
# rlang::abort('either use radii or radius1, radius2')
# }
#
# if (missing(radii)) {
# radii <- c(radius1, radius2)
# }
# bestd <- optimize(opt_d, lower = abs(diff(radii)), upper = sum(radii),
# area = area, radii = radii)
# }
To get that to work with a mutate, we need to feed it both radii. we could do that long or wide, but given that we usually have long data:
Make a column identifying the set, another with the set intersection, and use that to make the d in a grouped mutate.
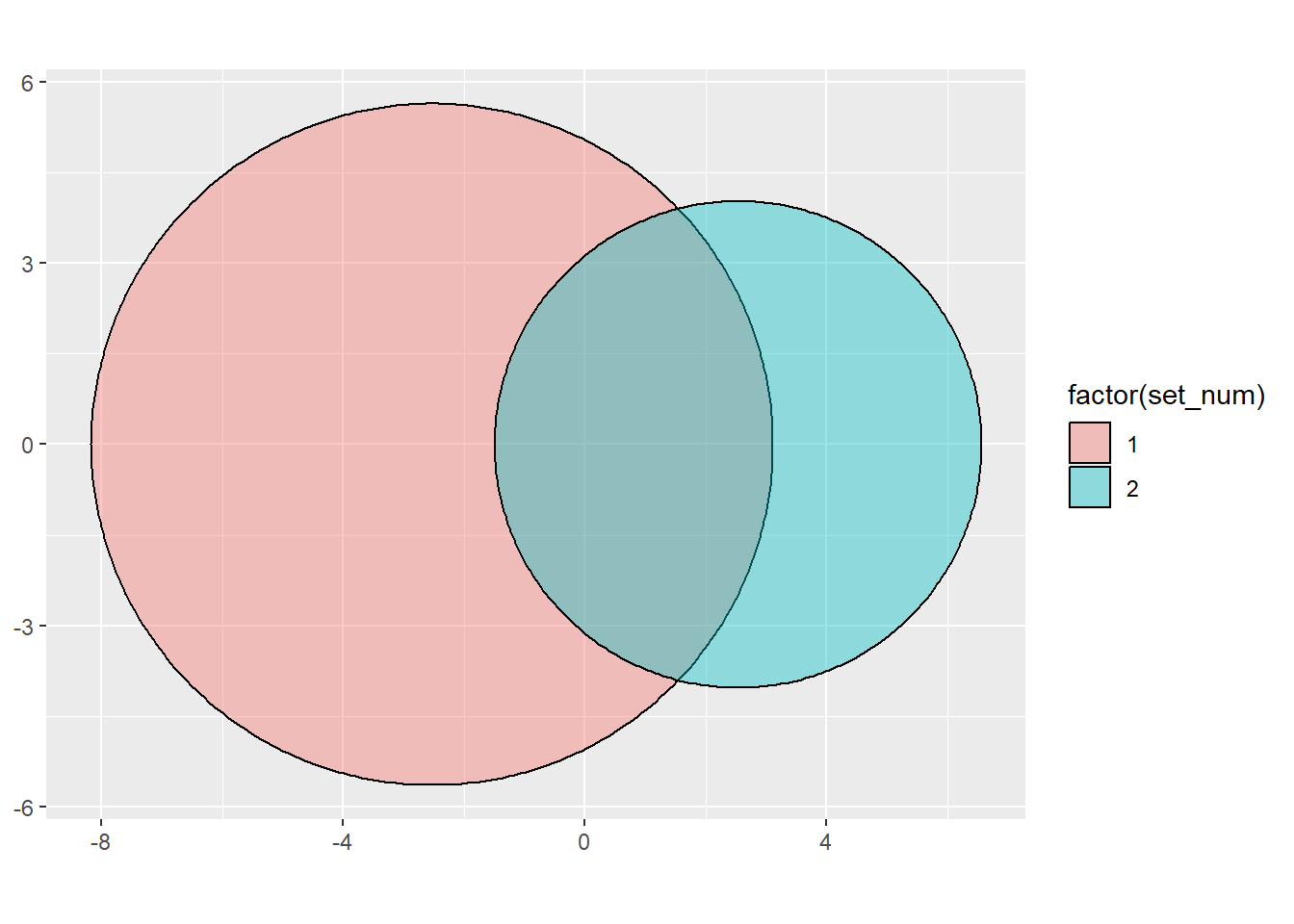
That really is fairly contrived to have the separate circles long. But when it comes time to plot, it’s WAY nicer.
Let’s say we have a dataset where we have already calculated the sizes and overlaps of a bunch of sets. Just make it long, since that’s how the data we want to use will be, and it makes the plotting easier.
Now, to make the circles, we need to calculate radii and distance between centers. This is now equally as annoyign to be wide
manysets <- manysets |>
mutate(radius = sqrt(area/pi)) |>
mutate(d = find_d(unique(overlap), radii = radius), .by = set_pair)
# makes testing easier
manysets <- manysets |>
# and a check
mutate(area_calc = calc_a(unique(d), radii = radius), .by = set_pair) |>
mutate(area_error = abs(overlap - area_calc)) |>
mutate(centers = d*c(-0.5, 0.5))Now to plot
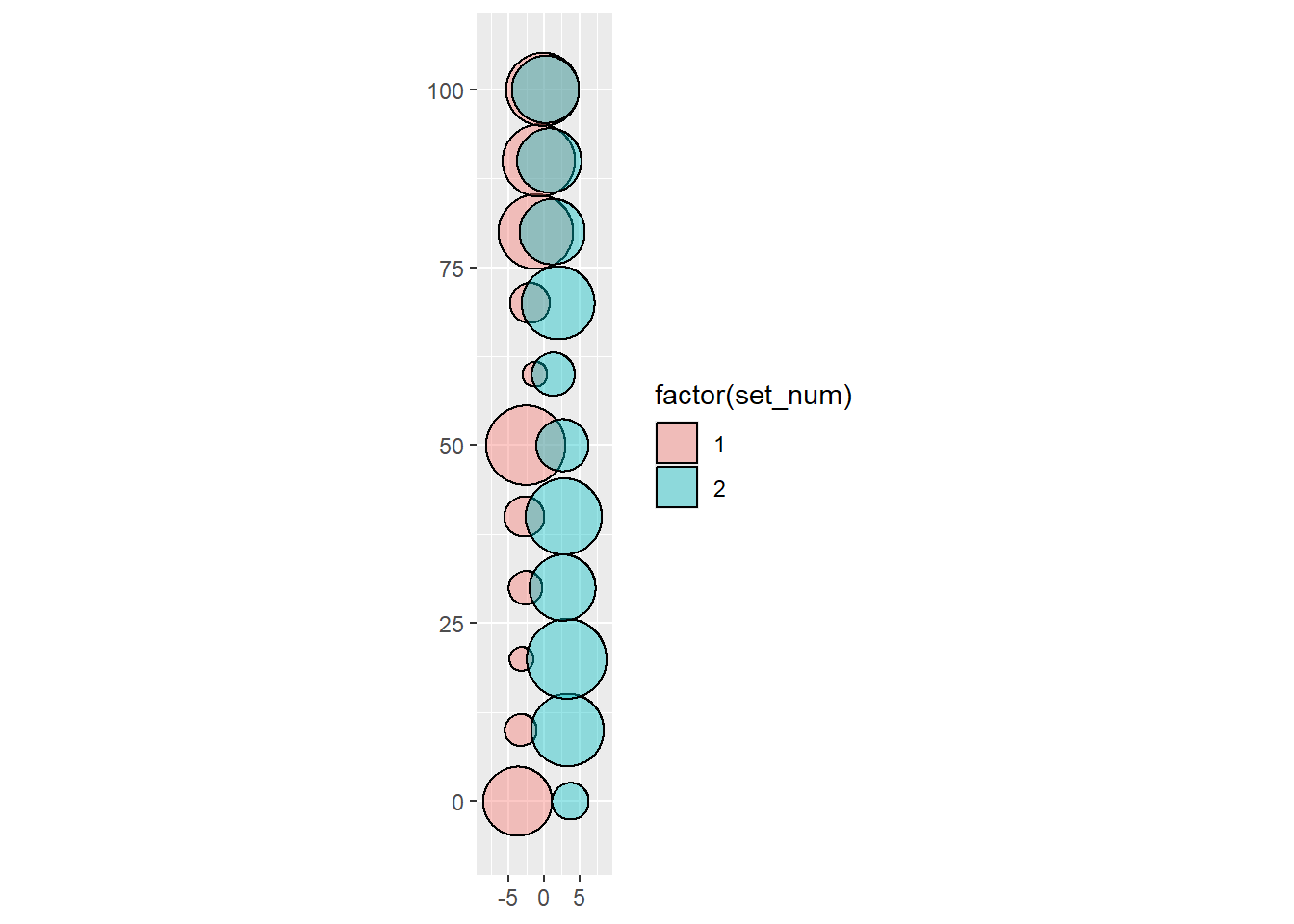
And the area_error is always very low
# A tibble: 22 × 10
set_pair set_num area overlapfrac overlap radius d area_calc area_error
<int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 73.7 0 0 4.84 7.42 0.00000176 0.00000176
2 1 2 20.8 0 0 2.58 7.42 0.00000176 0.00000176
3 2 1 15.8 0.1 1.58 2.24 6.56 1.58 0.0000554
4 2 2 81.7 0.1 1.58 5.10 6.56 1.58 0.0000554
5 3 1 8.83 0.2 1.77 1.68 6.36 1.77 0.0000764
6 3 2 98.5 0.2 1.77 5.60 6.36 1.77 0.0000764
7 4 1 17.3 0.3 5.20 2.35 5.21 5.20 0.0000348
8 4 2 67.5 0.3 5.20 4.63 5.21 5.20 0.0000348
9 5 1 24.9 0.4 9.96 2.81 5.55 9.96 0.0000984
10 5 2 89.7 0.4 9.96 5.34 5.55 9.96 0.0000984
# ℹ 12 more rows
# ℹ 1 more variable: centers <dbl>The way this work is probably easier to see not with random sizes, but consistent
Now get r, d, and centers
evensets <- evensets |>
mutate(radius = sqrt(area/pi)) |>
mutate(d = find_d(unique(overlap), radii = radius), .by = set_pair) |>
mutate(centers = d*c(-0.5, 0.5))
# makes testing easier
evensets <- evensets |>
# and a check
mutate(area_calc = calc_a(unique(d), radii = radius), .by = set_pair) |>
mutate(area_error = abs(overlap - area_calc))Now to plot
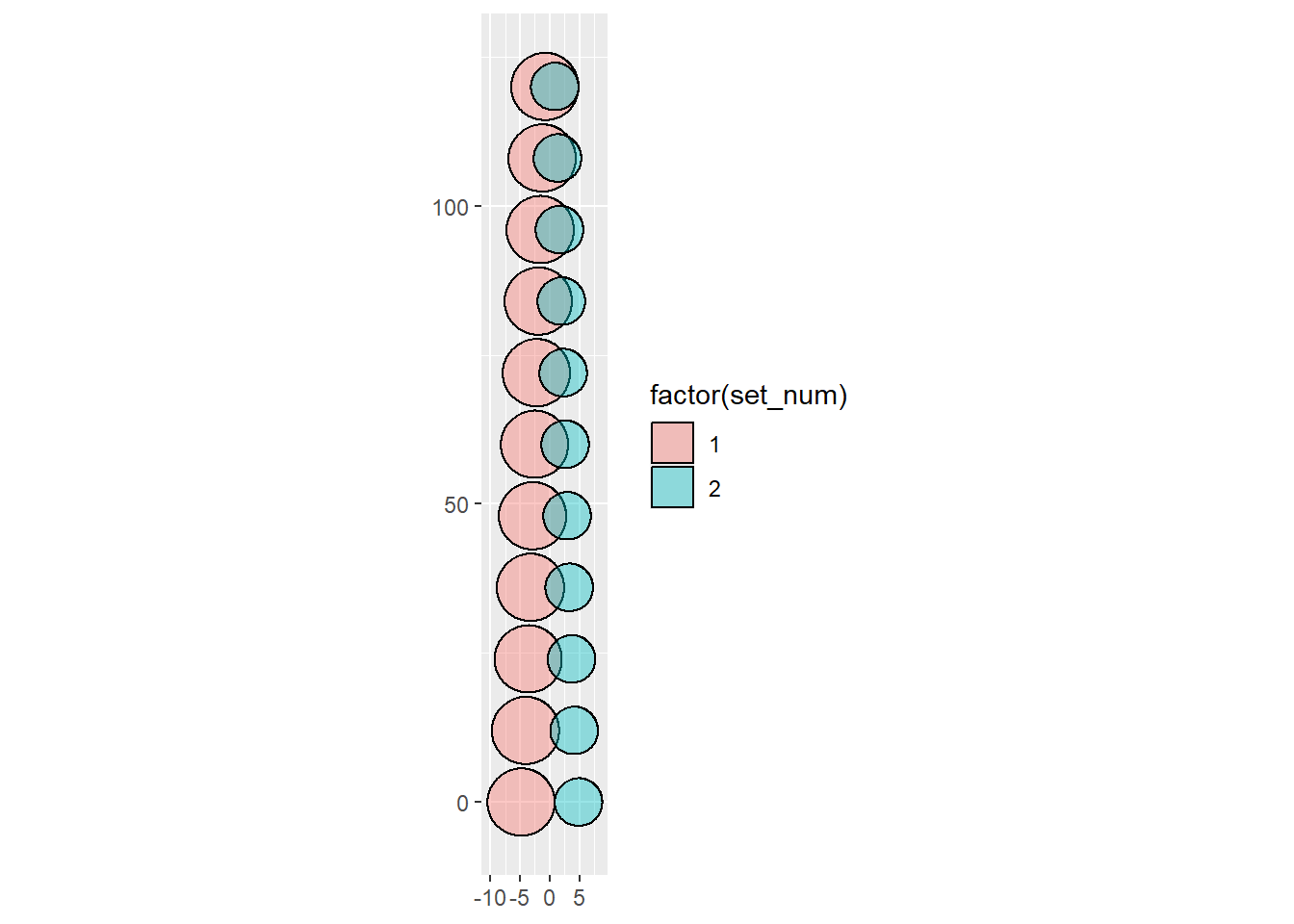
One thing I’m going to want to do when I use this is scale them (e.g. I’ll have massive numbers, but need to scale them down). I think the easiest way to do that is to just operate on scaled areas and overlaps.
So, recapitulating the above but with larger numers and then scaled
Now get r, d, and centers
scaledsets <- scaledsets |>
mutate(radius = sqrt(area/pi),
radius_scaled = sqrt(area_scaled/pi)) |>
mutate(d = find_d(unique(overlap), radii = radius),
d_scaled = find_d(unique(overlap_scaled), radii = radius_scaled),
.by = set_pair) |>
mutate(centers = d*c(-0.5, 0.5),
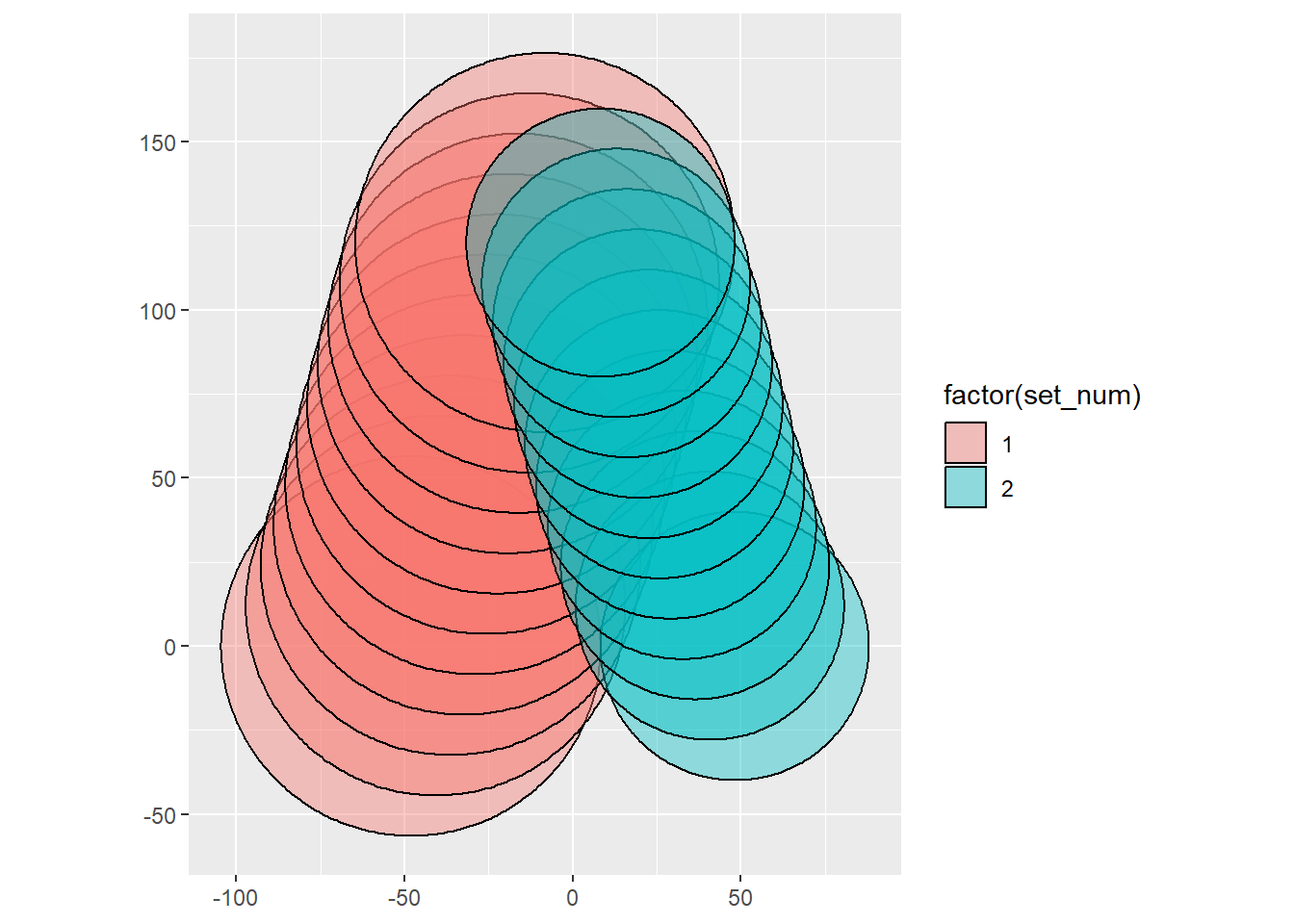
centers_scaled = d_scaled*c(-0.5, 0.5))Now to plot First the raw
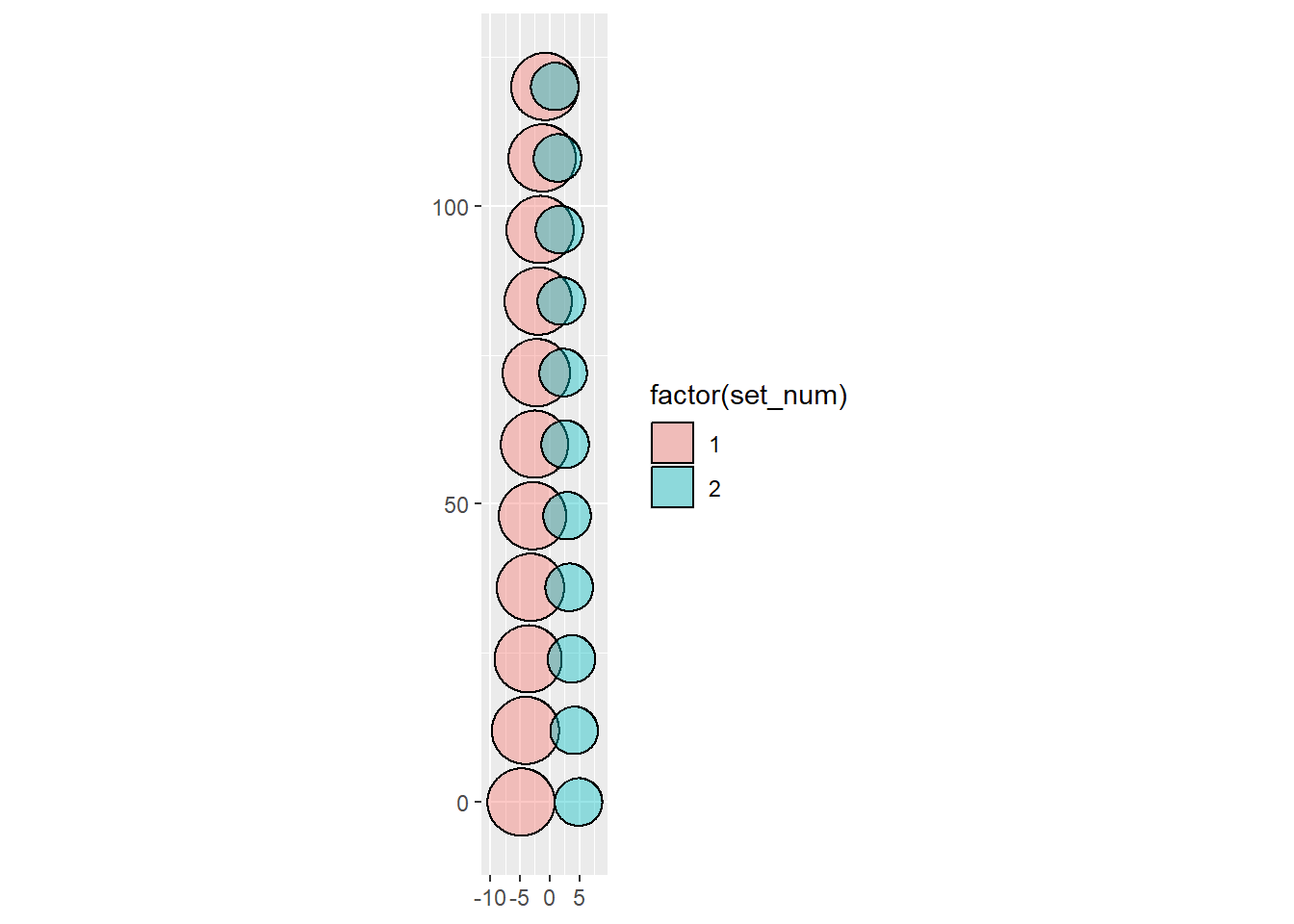
Then the scaled, looks like before.
Now, if the goal is just to plot, can I (should I) write a little function that just returns radii and centers? It would require a very specific data format, so not sure it’s worth it. Obviously it could be generalised to long, wide, and as here, partially-long (areas in col, but overlap in another). I think I might see how useful it would be when I actually try to do this for my data and then decide
We could also use d to go in any direction, with any offset. E.g. from a base 0,0, we could shift only one circle right or left or up or down or given an angle and pythagoras off at an arbitrary angle.